What happens if you publicly release more than 10.000 biopsies to challenge AI developers around the world? Is AI for pathology ready for clinical implementation? How powerful are challenges in catalyzing new AI developments? Do algorithms trained on EU data and reference standards generalize to US settings? Can you compare algorithms across different continents and reference standards? In our latest research, now published in Nature Medicine, we aimed to address these questions.
Introduction
Artificial intelligence (AI) has shown promise for diagnosing prostate cancer in biopsies.1,2,3,4,5 For example, in my Ph.D. research, we have shown that AI can grade prostate cancer at the level of experienced pathologists and can also support them during their review. However, results have generally been limited to individual studies, lacking validation in multinational settings. Competitions (“Challenges”) have been shown to be accelerators for medical imaging innovations, with the CAMELYON challenge being a prime example. Though, the impact of challenges is often hindered by a lack of reproducibility and independent validation.
For the 1.3 million new prostate cancer patients yearly, the Gleason grade of their biopsies is a crucial element for treatment planning. Pathologists characterize tumors into different Gleason growth patterns based on the histologic architecture of the tumor tissue. Based on the distribution of Gleason patterns, biopsy specimens are categorized into one of five groups, with a higher number denoting a worse outlook for the patient. Unfortunately, this process is subjective as pathologists frequently differ in their assessments. This could have significant consequences for a patient, for example leading to undergrading and overgrading of prostate cancer.
With a large team from Radboud University Medical Center, Karolinska Institute, Tampere University, Google Health, and Kaggle, we have organized the PANDA challenge: Prostate cANcer graDe Assessment using the Gleason grading system. We aimed to set the next step in automated Gleason grading for prostate cancer with this new challenge.
For the PANDA challenge, we released all training data of two major studies4,5 on automated Gleason grading, both previously published in Lancet Oncology. The training set contained almost 11.000 prostate biopsies, with slide-level labels and label masks. During the competition, 1010 teams with a total of 1290 developers from 65 countries joined the challenge. With these numbers, the PANDA challenge is, to the best of our knowledge, the largest challenge organized for pathology to date.
The challenge results are now available online in our paper AI for diagnosis and Gleason grading of prostate cancer: the PANDA challenge as published in Nature Medicine.

With more than 10.000 images for training and over one thousands participants, the PANDA challenge was the largest challenge organized for pathology to date.
Setup of the challenge
The main goal for all challenge participants was to design an algorithm that could automatically assign a Gleason grade group to a biopsy specimen. There was no limitation on the chosen methods. Still, teams had to follow some instructions to make sure they could correctly load the biopsies during evaluations.
Unique to the PANDA challenge was the way the study was set up. We wanted to make sure there was no room for “cheating,” and that results would be translatable to other datasets and settings. In many previous challenges, test data is released, and participants needed to submit their algorithms predictions to a central system. Even with the ground truth labels hidden, there is a risk that teams will tune their algorithm to the test data or even hand labeling the data themselves. Therefore, we asked teams to submit their algorithms instead. The test data was kept private at all times.
Throughout the competition, teams could request evaluations of their algorithm on the public leader board. This was done by submitting a new version of their algorithm. The algorithms were then simultaneously blindly validated on the private set. The algorithms were required to analyze 1000 biopsies within 6 hours for this process, but most algorithms needed less time than that.
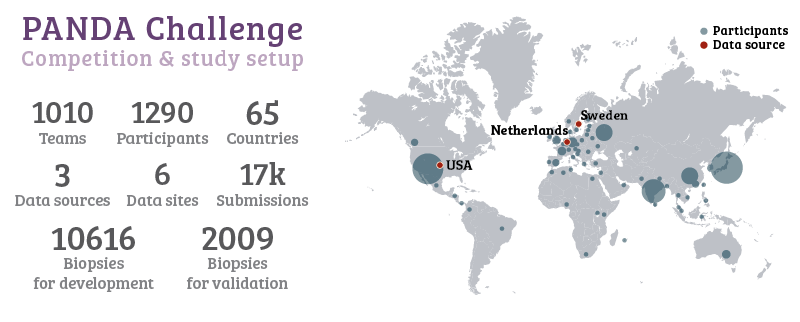
More than a thousand developers from 65 different countries joined the challenge.
After the competition ended, we selected fifteen teams to join for extensive independent validation of their algorithms on new data. The selection was based on the score on the final leaderboard and method description, and scientific contribution. The latter criteria were used to ensure we had a good representation of algorithms for the analysis.
We reproduced these fifteen algorithms fully in separate cloud systems without the original developers. This made sure we evaluated the algorithms as-is, without any additional tuning. We then applied the algorithms to the new data from the EU5 and the US2. The challenge, combined with these additional evaluations, resulted in a total of 32.137.756 biopsies processed by the algorithms. As with the public/private leaderboard data, we kept the data from the extended validations secret. In fact, the teams did not even know that their algorithms would be analyzed on this data when they designed them. We tried to simulate “AI in the wild” with this setup, as in real-life scenarios, algorithms will also be applied to completely unseen and new data.
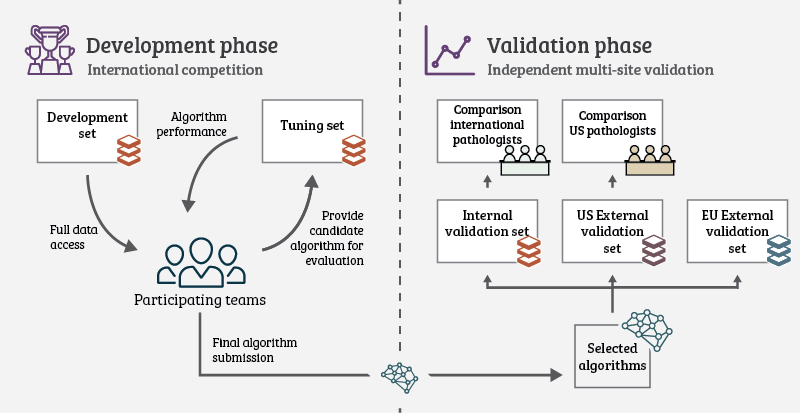
Setup of the challenge was split in two phases: A competition phase (the actual challenge), and a validation phase where the top algorithms were evaluated on new data.
Crowdsourced AI
Challenges are often a powerful way of crowdsourcing new AI innovations. The PANDA challenge was no exception: Due to the scale of the competition, after ten days, one of the algorithms was already at the level of the average pathologist. In the remainder of the challenge, many teams caught up and improved further. This speed in development was also driven by extensive sharing of tips and tricks through the challenge forums.
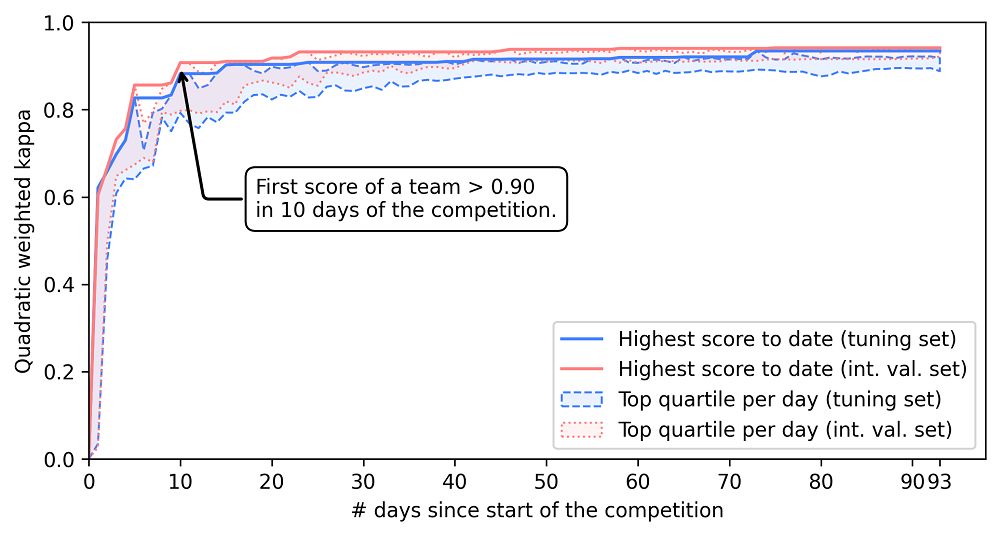
Within ten days of the competition the first agreement achieved pathologist-level performance (defined as agreement of 0.9 or higher with the reference standard.
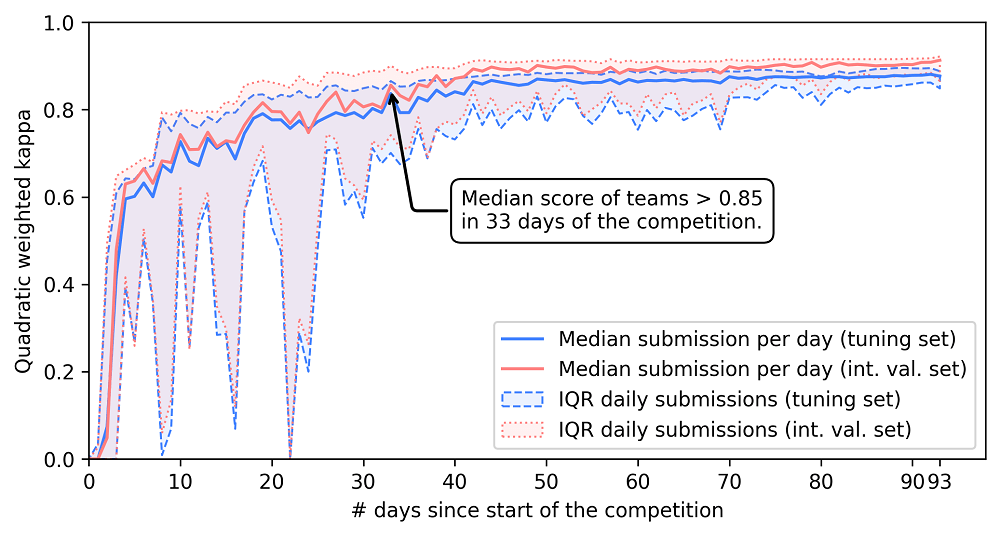
Due to intensive discussions on the challenge forum on how to best approach the problem, many teams quickly achieved high performing algorithms.
A complete discussion of all results is out of the scope of this blog post; for that, I recommend checking our paper, which is Open Access. In the paper, we compare the algorithms on several datasets and two pathologists panels. We also dive deeper into the question of whether it’s actually feasible to evaluate algorithms across datasets with different reference standards.
Our main results from the validation phase can be summarized as follows: We found that the AI algorithms successfully generalized across different patient populations, laboratories, and reference standards. Although only trained using data from EU pathologists and never tuned to new data, the algorithms retained their performance compared to pathologists from the US. This is an exciting result as it shows that AI is ready for clinical implementation.
In the paper we report more results on all datasets and show how these algorithms hold up compared to different panels of pathologists.
Acessing the PANDA dataset
We made the entire training set of 10,616 digitized de-identified H&E stained prostate biopsies (383GB) publicly available for further non-commercial research. The data can be used under a Creative Commons BY-SA-NC 4.0 license. To adhere to the “Attribution” part of the license, we ask anyone who uses the data to cite the corresponding paper (see Acknowledgements). In addition, you can also cite the two papers4,5 that describe the original data collection.
The latest information about downloading and using the PANDA dataset is available on the PANDA Challenge website. For now, the quickest way to access the data is through the competition on Kaggle.
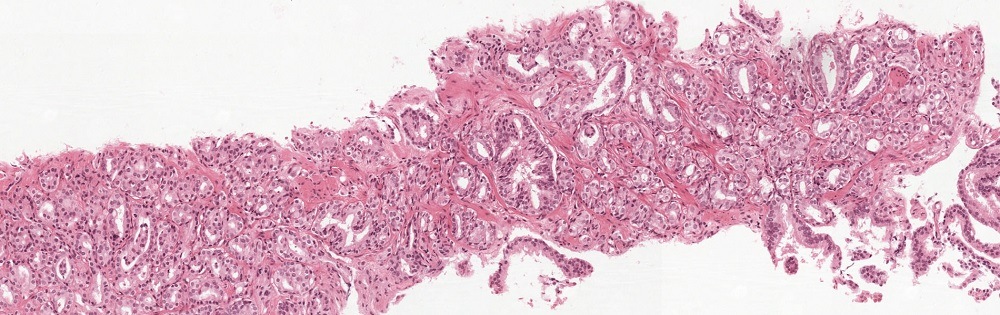
Using the PANDA dataset
To quickly get started with the PANDA dataset, we made a Jupyter notebook with some examples. You can find this notebook in our Github repository or try it out directly on Kaggle.
If you want a quick start, you can read a patch from one of the slides with a few lines of Python and OpenSlide:
import os
import openslide
from IPython.display import Image, display
# Open the image (does not yet read the image into memory)
image = openslide.OpenSlide(os.path.join(data_dir, '005e66f06bce9c2e49142536caf2f6ee.tiff'))
# Read a specific region of the image starting at the upper left coordinate (x=17800, y=19500) on level 0 and extract a 256*256 pixel patch.
# At this point image data is read from the file and loaded into memory.
patch = image.read_region((17800,19500), 0, (256, 256))
# Display the image
display(patch)
# Close the opened slide after use
image.close()
After running the snippet above, you should see the following (a single patch):
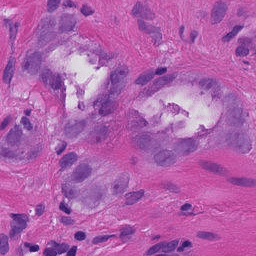
Want to try a late submission?
It’s still possible to participate in the challenge and evaluate your algorithm on the internal validation set. Please go to the competition page on Kaggle for more information about these late submissions. Note that you will need to submit a working algorithm to receive a score on the test set.
Acknowledgements
What started with a small email in 2019 ended up in a vast project that spanned over two years. The PANDA challenge has been a group effort by researchers, pathologists, technicians, developers, and many others.
Author list: Wouter Bulten, Kimmo Kartasalo, Po-Hsuan Cameron Chen, Peter Ström, Hans Pinckaers, Kunal Nagpal, Yuannan Cai, David F. Steiner, Hester van Boven, Robert Vink, Christina Hulsbergen-van de Kaa, Jeroen van der Laak, Mahul B. Amin, Andrew J. Evans, Theodorus van der Kwast, Robert Allan, Peter A. Humphrey, Henrik Grönberg, Hemamali Samaratunga, Brett Delahunt, Toyonori Tsuzuki, Tomi Häkkinen, Lars Egevad, Maggie Demkin, Sohier Dane, Fraser Tan, Masi Valkonen, Greg S. Corrado, Lily Peng, Craig H. Mermel, Pekka Ruusuvuori, Geert Litjens, Martin Eklund and the PANDA Challenge consortium.
Please use the following to refer to our paper or this post:
Bulten, W., Kartasalo, K., Chen, PH.C. et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat Med (2022). https://doi.org/10.1038/s41591-021-01620-2
@article{Bulten2022,
doi = {10.1038/s41591-021-01620-2},
url = {https://doi.org/10.1038/s41591-021-01620-2},
year = {2022},
month = jan,
publisher = {Springer Science and Business Media {LLC}},
author = {Wouter Bulten and Kimmo Kartasalo and Po-Hsuan Cameron Chen and Peter Str\"{o}m and Hans Pinckaers and Kunal Nagpal and Yuannan Cai and David F. Steiner and Hester van Boven and Robert Vink and Christina Hulsbergen-van de Kaa and Jeroen van der Laak and Mahul B. Amin and Andrew J. Evans and Theodorus van der Kwast and Robert Allan and Peter A. Humphrey and Henrik Gr\"{o}nberg and Hemamali Samaratunga and Brett Delahunt and Toyonori Tsuzuki and Tomi H\"{a}kkinen and Lars Egevad and Maggie Demkin and Sohier Dane and Fraser Tan and Masi Valkonen and Greg S. Corrado and Lily Peng and Craig H. Mermel and Pekka Ruusuvuori and Geert Litjens and Martin Eklund and {The PANDA challenge consortium}},
title = {Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the {PANDA} challenge},
journal = {Nature Medicine}
}
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
Improve prostate cancer diagnosis: participate in the PANDA Gleason grading challenge
Can you build a deep learning model that can accurately grade protate biopsies? Participate in the PANDA challenge
The potential of AI in medicine: AI-assistance improves prostate cancer grading
In a completely new study we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
Epithelium segmentation using deep learning and immunohistochemistry
We developed a new deep learning method to segment epithelial tissue in digitized hematoxylin and eosin (H&E) stained prostatectomy slides using immunohistochemistry (IHC) as reference standard.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.
References
The potential of AI in medicine: AI-assistance improves prostate cancer grading ↩︎
Steiner, D. F. et al. Evaluation of the Use of Combined Artificial Intelligence and Pathologist Assessment to Review and Grade Prostate Biopsies. JAMA Network Open 3, e2023267 (2020). Read online ↩︎ ↩︎
Bulten, W. et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Modern Pathology (2020) Read online ↩︎
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. The Lancet Oncology 21, 233-241, Read online (2020). ↩︎ ↩︎ ↩︎
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. The Lancet Oncology 21, 222-232, Read online (2020). ↩︎ ↩︎ ↩︎ ↩︎
