To improve prostate cancer detection and grading algorithms, we required a system that could precisely outline epithelial tissue. Our main idea was that such a system could automatically refine coarse annotations made by human annotators or other deep learning systems; for example in our project on automated Gleason grading.
At first, we trained a system using the conventional way: based on human annotations, we trained a U-Net in a simple patch-based approach. Unfortunately, we were hindered by time and the limits of human performance: the system’s performance can at most be as best as the annotations of the data. In the case of prostate cancer, epithelial tissue can express as individual cells laying in groups in the stroma, which makes manual annotating data a time-consuming and challenging task. With this project, we set out to develop a novel method to circumvent the need for elaborate manual annotations.
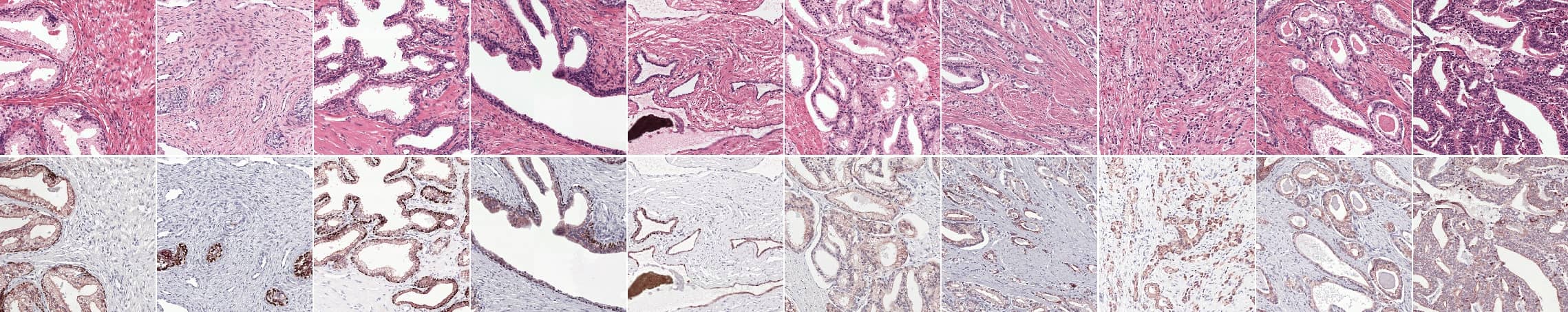
Prostate epithelial tissue can express itself in many forms. The first four columns show examples of benign tissue, the last four of prostate cancer. Top row shows the original H&E, the bottom row IHC.
Data (the PESO dataset)
We have developed our system using a new dataset of 102 prostatectomy tissue blocks. From each block a new section was cut, stained with H&E and scanned. After scanning, the tissue was destained, restained using immunohistochemistry, and scanned again. All slides were scanned at 20x magnification (pixel resolution 0.24 μm).
We used two markers for the immunohistochemistry: CK8/18 (using DAB) to mark all glandular epithelial tissue (benign and malignant), and P63 (using NovaRED) for the basal cell layer, which is normally present in benign glands but not in malignant glands. Restaining, instead of making consecutive slides, results in an H&E and IHC whole-slide image (WSI) pair for each patient that contains the same tissue.
We have released the H&E images used in this project as a public dataset on Zenodo: the PESO dataset. More information on this dataset and how to use it can be found in a separate blog post.
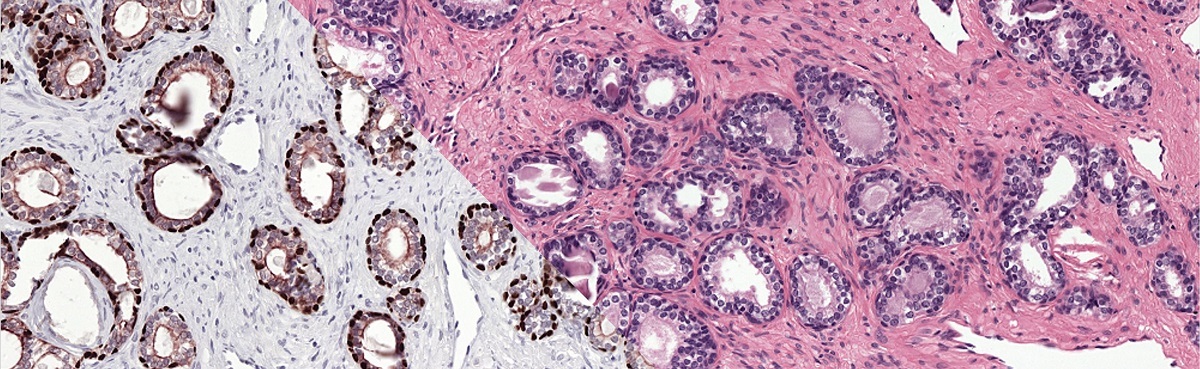
To train our system we make use of registered H&E and IHC stained slides. Registration makes it possible to transfer annotations from one domain to the other.
Method
To train our system, we used a two-step approach. First, we trained a convolutional network to segment epithelium in the IHC slides. By applying color deconvolution and subsequent recognition of positively stained pixels, we were able to have extensive training data while preventing the cumbersome and imprecise process of manually annotating epithelial regions. After this first step, we transfered the annotations to the H&E slides and trained the final network. The steps are described in more detail below:
- On a subset of the IHC dataset, we applied color deconvolution to select the brown color channel. Some binary operations were used to remove small artifacts. The resulting mask contains most of the epithelial tissue but also includes non epithelial tissue that was colored by the stain (such as corpora amylacea).
- On this subset, we corrected significant errors by hand. Artifacts were marked as such. Note that this task is only a fraction of the work; instead of annotating individual epithelial cells, only errors have to be annotated in a subset of the slides.
- We trained a first U-Net on the corrected IHC slides. This network could then be used to generate epithelial masks for all the IHC slides (including the non-corrected slides).
- We registered the H&E and IHC slides. As the original tissue was restained, the masks generated by the IHC network matched the H&E slides perfectly.
- The final network was trained using the transferred masks.
All hyperparameters for both U-Nets can be found in the paper.
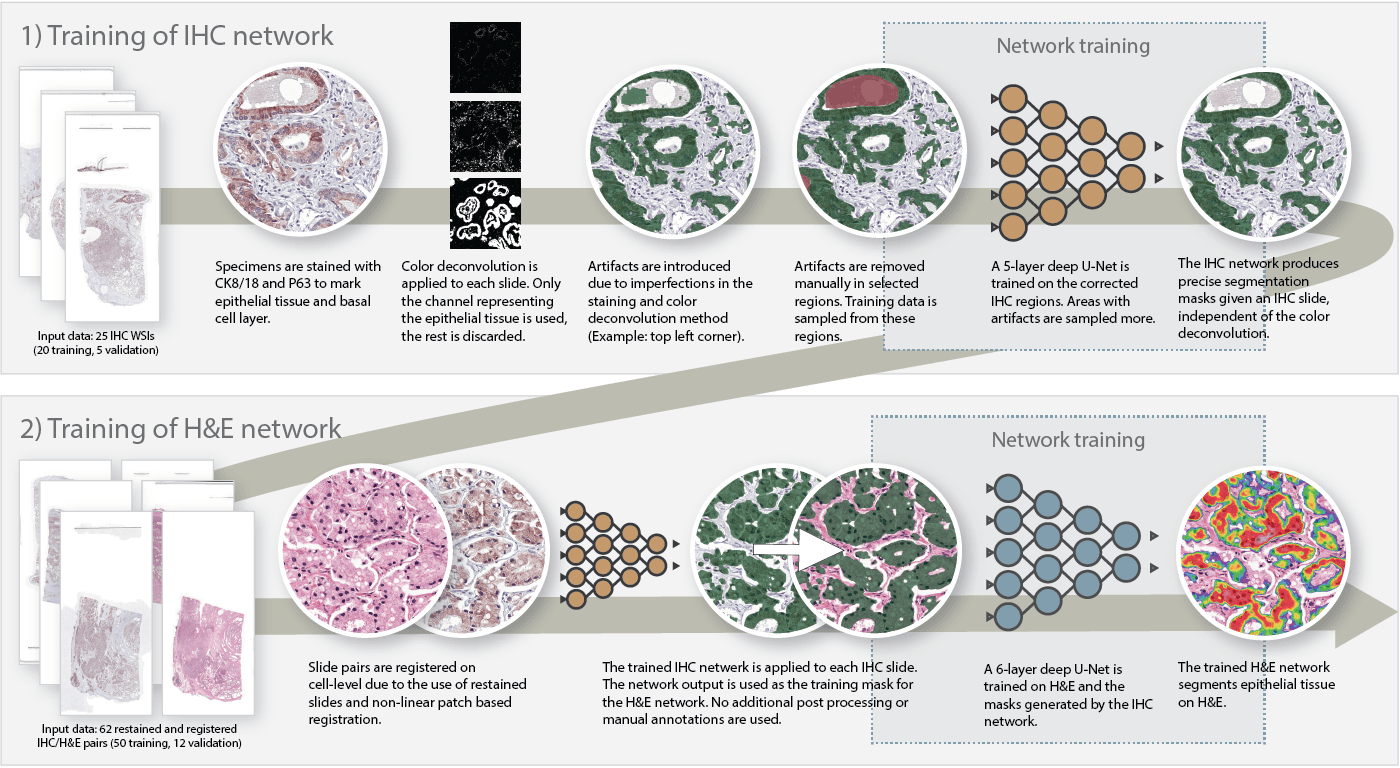
Method used to segment epithelial tissue. First we train a system to segment epithelial tissue on IHC, later we transfer this to H&E to train the final system.
Results
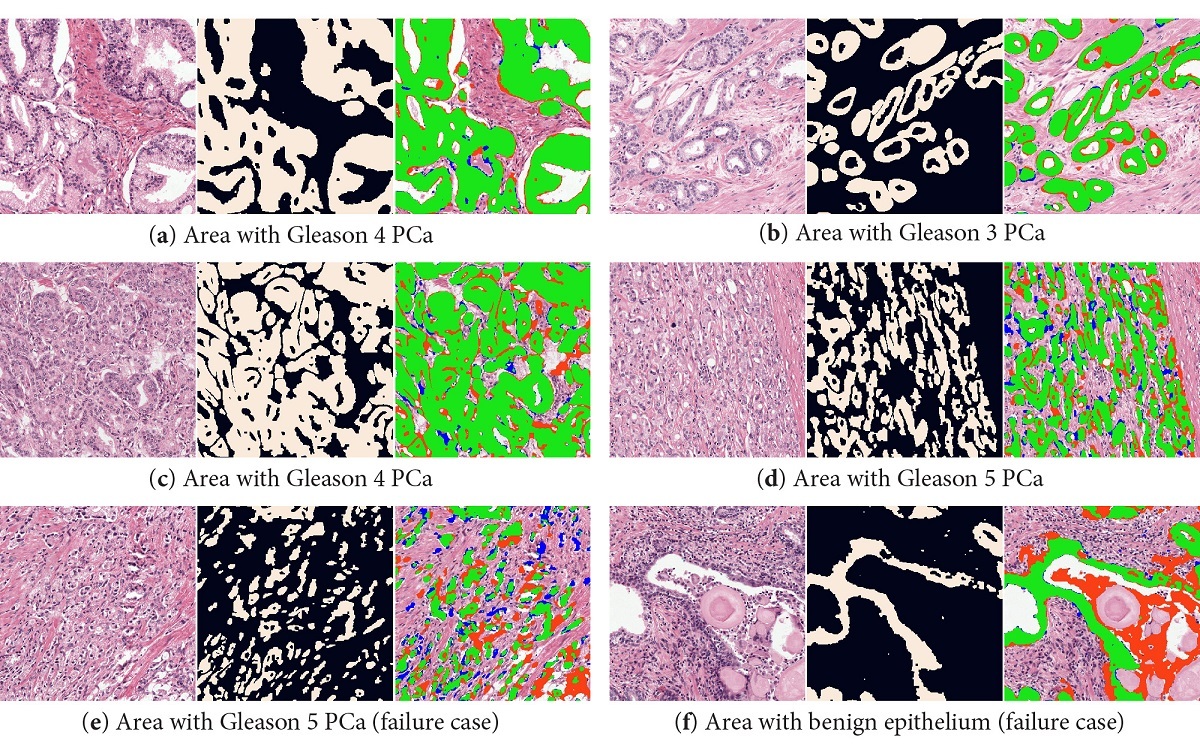
Our system was able to accurately segment epithelial tissue, both in benign and cancerous regions (overall F1 score of 0.893). Even in regions with high grade prostate cancer, the system is able to segment individual cells. Some problems occurred in regions with high inflammation (that can look very similar to epithelial tissue). Correcting the color deconvolution masks increased the performance, but even more important removed consistent misclassifications of non-epithelial regions (like corpora amylacea). For a complete overview of all results, including results on an external dataset, please refer to the paper (Open Access).
The final U-Net was a critical component in the automatic data labeling technique of our project on automated Gleason grading. Using the epithelial masks, we were able to generate precise gland-level outlines of benign and tumorous tissue in prostate biopsies.
More info
Read full paper on Scientific Reports
This work was financed by a grant from the Dutch Cancer Society (KWF). You can use the following reference if you want to cite the paper:
Bulten, Wouter, et al. “Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard.” Scientific reports 9.1 (2019): 864.
Or, if you prefer BibTeX:
@article{bulten2019epithelium,
title={Epithelium segmentation using deep learning in H\&E-stained prostate specimens with immunohistochemistry as reference standard},
author={Bulten, Wouter and B{\'a}ndi, P{\'e}ter and Hoven, Jeffrey and van de Loo, Rob and Lotz, Johannes and Weiss, Nick and van der Laak, Jeroen and van Ginneken, Bram and Hulsbergen-van de Kaa, Christina and Litjens, Geert},
journal={Scientific reports},
volume={9},
number={1},
pages={864},
year={2019},
publisher={Nature Publishing Group}
}
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
AI for diagnosis of prostate cancer: the PANDA challenge
Artificial intelligence (AI) for prostate cancer analysis is ready for clinical implementation, shows a global programming competition, the PANDA challenge.
Improve prostate cancer diagnosis: participate in the PANDA Gleason grading challenge
Can you build a deep learning model that can accurately grade protate biopsies? Participate in the PANDA challenge
The potential of AI in medicine: AI-assistance improves prostate cancer grading
In a completely new study we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.


Lorenz Rumberger on
I really enjoyed the read and the actual paper. I'm really interested in the image registration pipeline that you used here, since I deal with a similar task. Are there more details on it or code somewhere available?