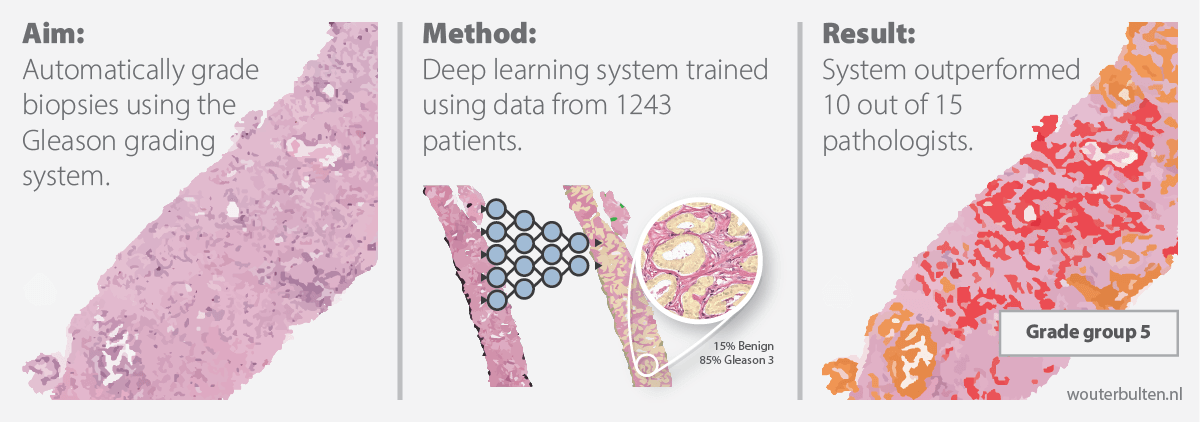
100-word summary: The Gleason score is the most important prognostic marker for prostate cancer patients but suffers from significant inter-observer variability. We developed a fully automated deep learning system to grade prostate biopsies. The system was developed using 5759 biopsies from 1243 patients. A semi-automatic labeling technique was used to circumvent the need for full manual annotation by pathologists. The developed system achieved a high agreement with the reference standard. In a separate observer experiment, the deep learning system outperformed 10 out of 15 pathologists. The system has the potential to improve prostate cancer prognostics by acting as a first or second reader.
Authors: Wouter Bulten, Hans Pinckaers, Hester van Boven, Robert Vink, Thomas de Bel, Bram van Ginneken, Jeroen van der Laak, Christina Hulsbergen-van de Kaa, Geert Litjens
Article URL: Paper at Lancet Oncology
arXiv URL: Preprint at arXiv
Automated Gleason Grading using Deep Learning and Artificial Intelligence.
Introduction
Prostate cancer is one of the most common forms of cancer, with more than 1.2 million new cases each year. The diagnosis of prostate cancer is complicated by multiple factors. Patients with low-grade prostate cancer are often better off with a wait-and-see approach than with active treatment due to the side effects of, for example, surgery. High-grade cancers, however, need to be diagnosed as soon as possible not to delay treatment and to increase patient survival. Unfortunately, diagnosis and grading of prostate cancer is a difficult task which suffers from inter- and Intra-observer variability. While export uropathologists have shown better concordance rates, such expertise is not available for every patient. In other words, there is a need for robust and reproducible grading at expert levels. We have developed an artificially intelligent system, using deep learning, that can perform the grading of prostate cancer at a pathologist-level performance.
In this blog post, we describe our approach, show how we tackled the problem of data labeling and, finally, show that our deep learning system operates at a pathologist-level performance. The article is now published at Lancet Oncology.
Background on Gleason grading
Treatment planning for prostate cancer patients is based mainly on the Gleason score of a biopsy. After the biopsy procedure, a pathologist examines the tissue through a microscope. Through the microscopic analysis, the pathologist needs to distinguish between benign and malignant tissue. Any malignant tissue is then classified according to the architectural pattern of the tumor. This classification system is formally described in the Gleason grading system.
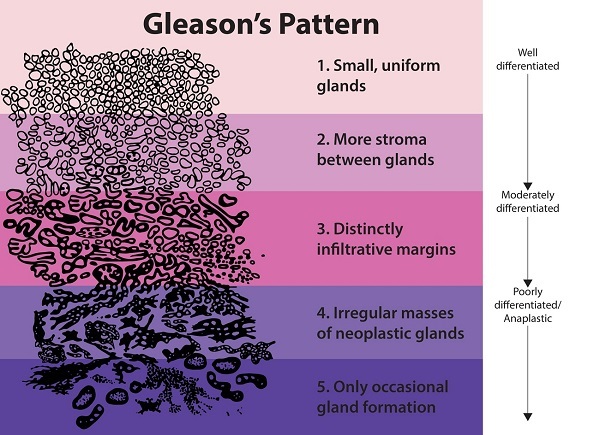
The Gleason scoring system.
In the Gleason grading system, a tumor region is assigned a number between 1 (low-risk) and 5 (high risk), though patterns 1 and 2 are not reported anymore for biopsies. For prostate biopsies, the most common pattern and the second-highest pattern together form the Gleason score, e.g., 3+5=8. To make reporting of prostate cancer more apparent for patients, Gleason scores are mapped to five prognostically different grade groups; with group 1 being the lowest risk, and 5 the highest.
Semi-automatic data labeling
Pathology archives are often full of tissue specimens, but detailed labels of these specimens are lacking. Deep learning, however, requires large annotated datasets to reach its full potential. Due to time and budget restrictions, it is often practically infeasible to hire pathologists to annotate the data.
In the case of Gleason grading, this problem is even more significant. High-grade cancers (Gleason growth pattern 5) can express in the form of (strands of) individual tumor cells. Individually outlining hundreds of these cells in thousands of tissue specimens is no entertaining endeavor.
We developed a novel approach to circumvent the need of manual annotations. First, we sampled cases from our dataset that contained only one Gleason grade (score 3+3, 4+4 or 5+5). Then we employed the following steps to label the data:
- For each case in our dataset, we used the original pathologist’s report (from diagnostics) to determine the Gleason score of a biopsy.
- A previously-trained tumor detection system was applied to all these biopsies. This procedure resulted in a rough outline of the tumor bulk. These tumor regions were still very coarse and needed further refinement. A description of this system can be found in the corresponding paper.
- The tumor masks were refined by filtering out all non-epithelial tissue. For this, we used a deep learning system that was trained using immunohistochemistry to detect epithelial cells. More information about this system can be read in the paper.
- All detected and filtered tumor cells were assigned with the Gleason grade of the biopsy.
- We trained an initial network on the automatically annotated data. After training, this segmentation network was able to assign Gleason growth patterns to individual cells.
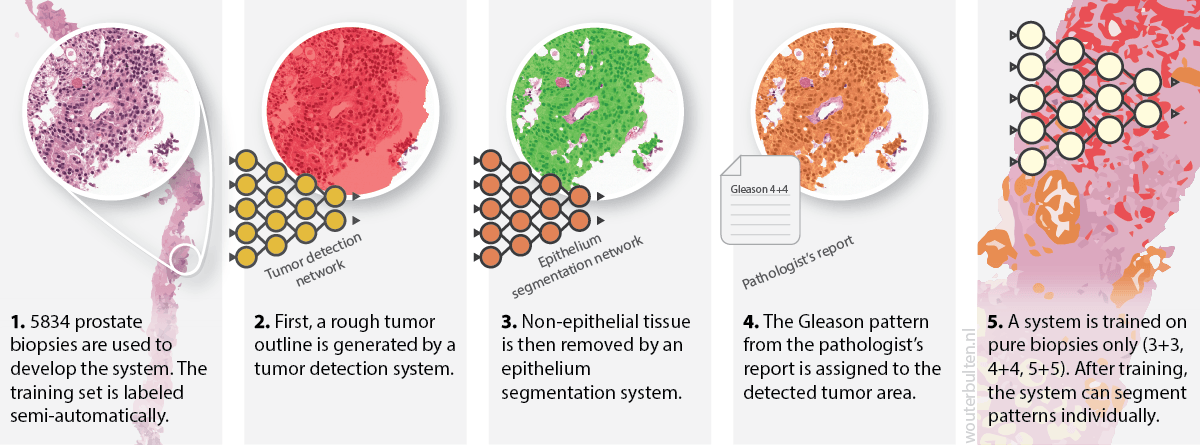
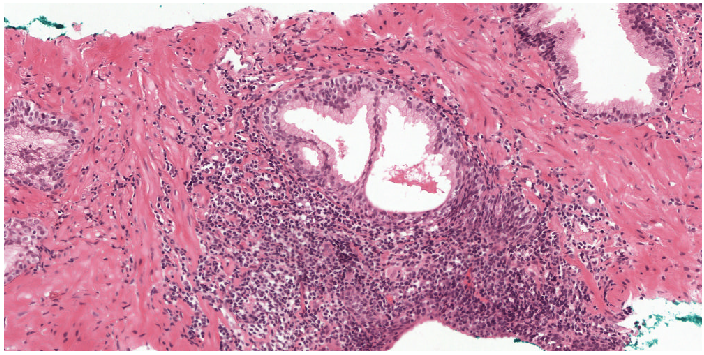
First part of our semi-automatic labeling method.
Network training
The trained network from step 5 (above) could then be used to annotate our complete dataset. This trained network was not perfect in assigning Gleason scores as it was only trained on a subset of our dataset. To annotate the full training set, we applied the network to all cases of our set. We then used the original pathologist’s report to fix any major mistakes. For example, a predicted Gleason 5 region in a biopsy with Gleason score 3+3 would be removed. Tissue that originated from benign biopsies, and was detected as malignant, was classified as “hard negative.”
With our full training set annotated, we could train the final deep learning system. The whole procedure required minimal human effort. Moreover, no pathologists were required to annotate the data; instead, we were able to utilize expert knowledge extracted from the pathologist’s reports. All training and network details can be found in the paper on arXiv.
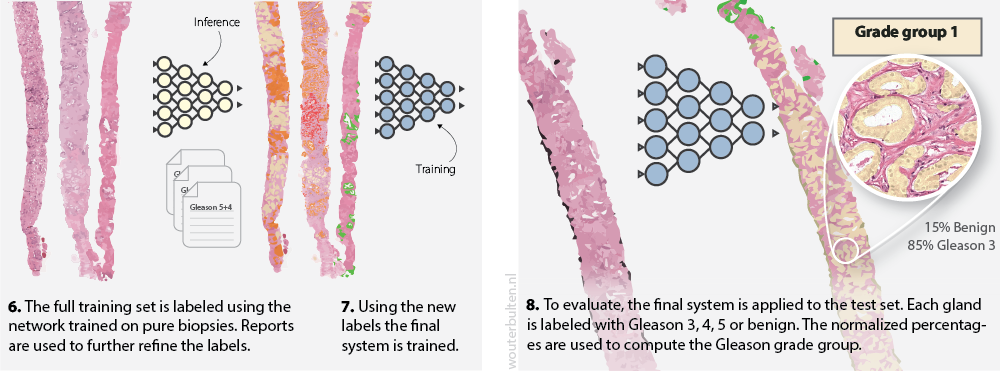
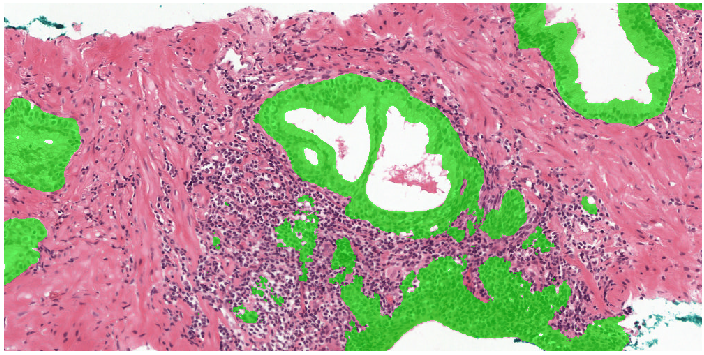
Second part of our semi-automatic labeling method and application of the system to the test set.
After the final network was trained, we were able to assign grade groups to biopsies automatically. Our system was trained as a segmentation network and outputted precise outlines of the tumor. Because of this segmentation, we can use a simple method to assign grade groups. Similar to clinical practice, we compute the volumes of each growth pattern. These volumes are then used to determine the primary and secondary Gleason grade, and finally, the grade group. We believe that this method is more interpretable and usable by pathologists in clinical practice. Each grade group prediction of the system can be easily checked by validating the marked glands.
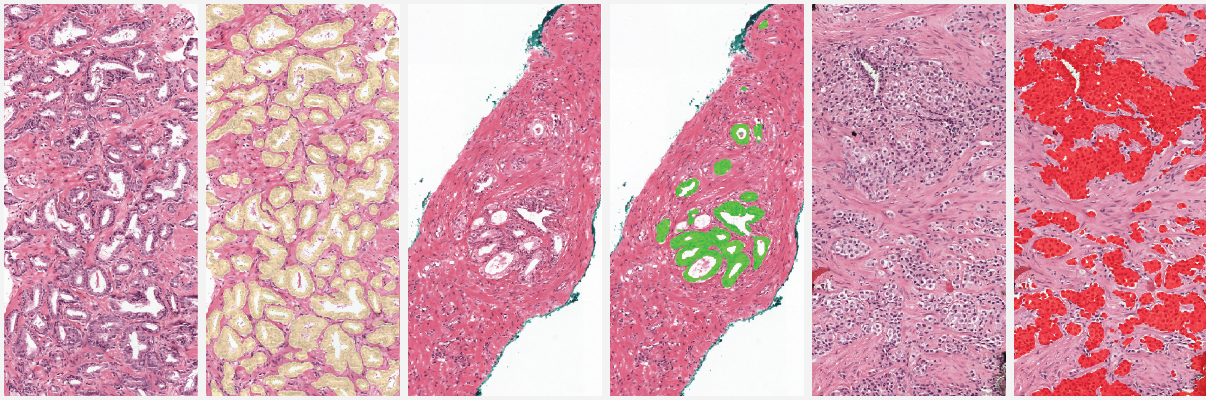
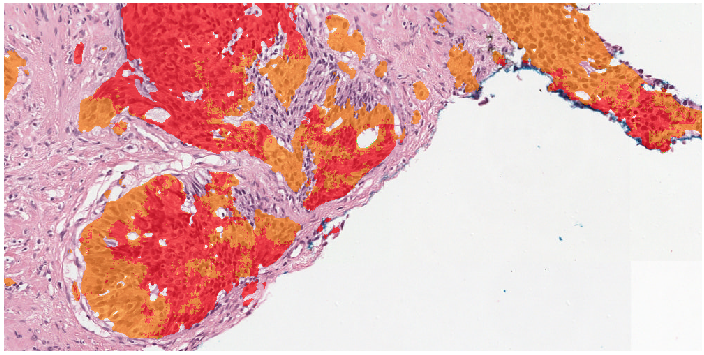
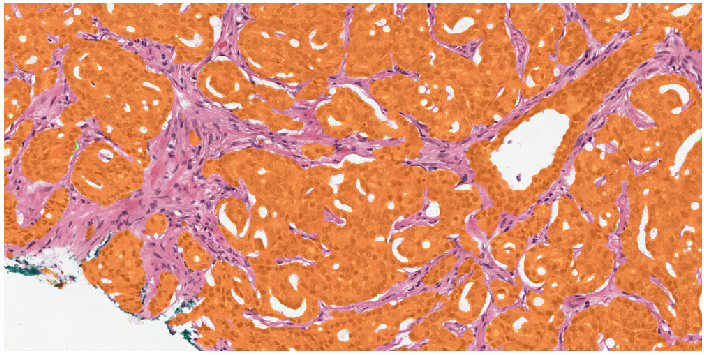
Example segmentation generated by the deep learning system: benign tissue (green), Gleason 3 (yellow), Gleason 5 (red).
Online examples
Two examples showing the raw output of the algorithm, without any post-processing, overlayed on biopsies. Benign tissue is colored green, Gleason 3 in yellow, Gleason 4 in orange and Gleason 5 in red. The original biopsy is shown on the left, the biopsy with overlay on the right. You can zoom in by scrolling, moving around can be done with click&drag.
Overview of main results
We compared our deep learning system on a test set of 535 biopsies. These biopsies were graded by three experts, and their consensus is used as the reference standard. The deep learning system achieved a high agreement with a quadratic Cohen’s kappa of 0.918.
Furthermore, from the test set, we selected 100 cases to be graded by an external panel. This panel consisted of 15 external raters (13 pathologists and 2 pathologists-in-training) from 10 different countries. In agreement with the reference standard (quadratic Cohen’s kappa) the deep learning system outperforms 10 out of 15 pathologists.
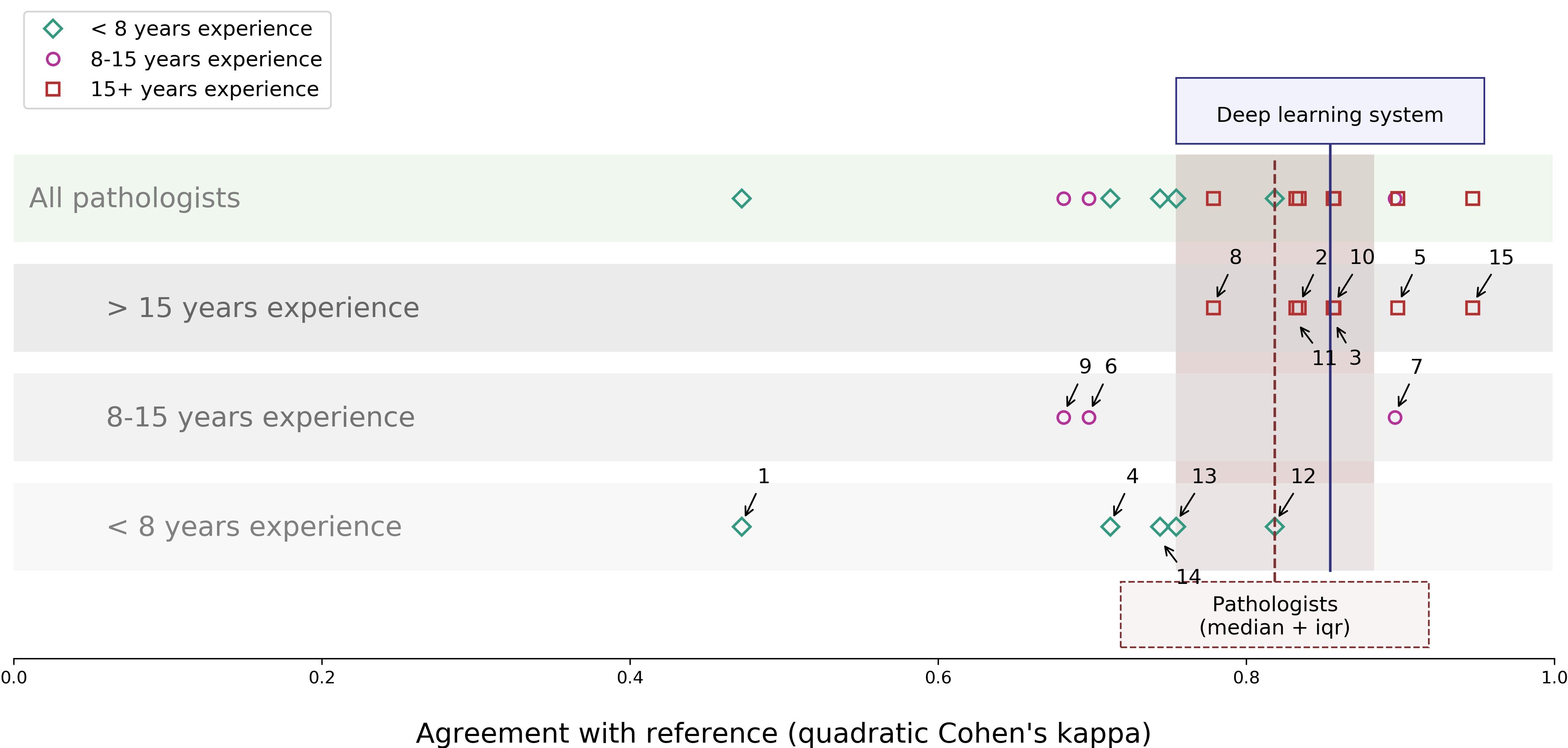
Comparison of the deep learning system with a panel of pathologists on agreement with the reference standard.
We also evaluated our deep learning system on grouping patients in prognostically relevant groups: (1) benign versus malignant biopsies; (2) using grade group 2 as cut-off; and (3) high versus low cancer. The deep learning system achieves a pathologist-level performance on all groups. Please see the full paper for further results and more detail.
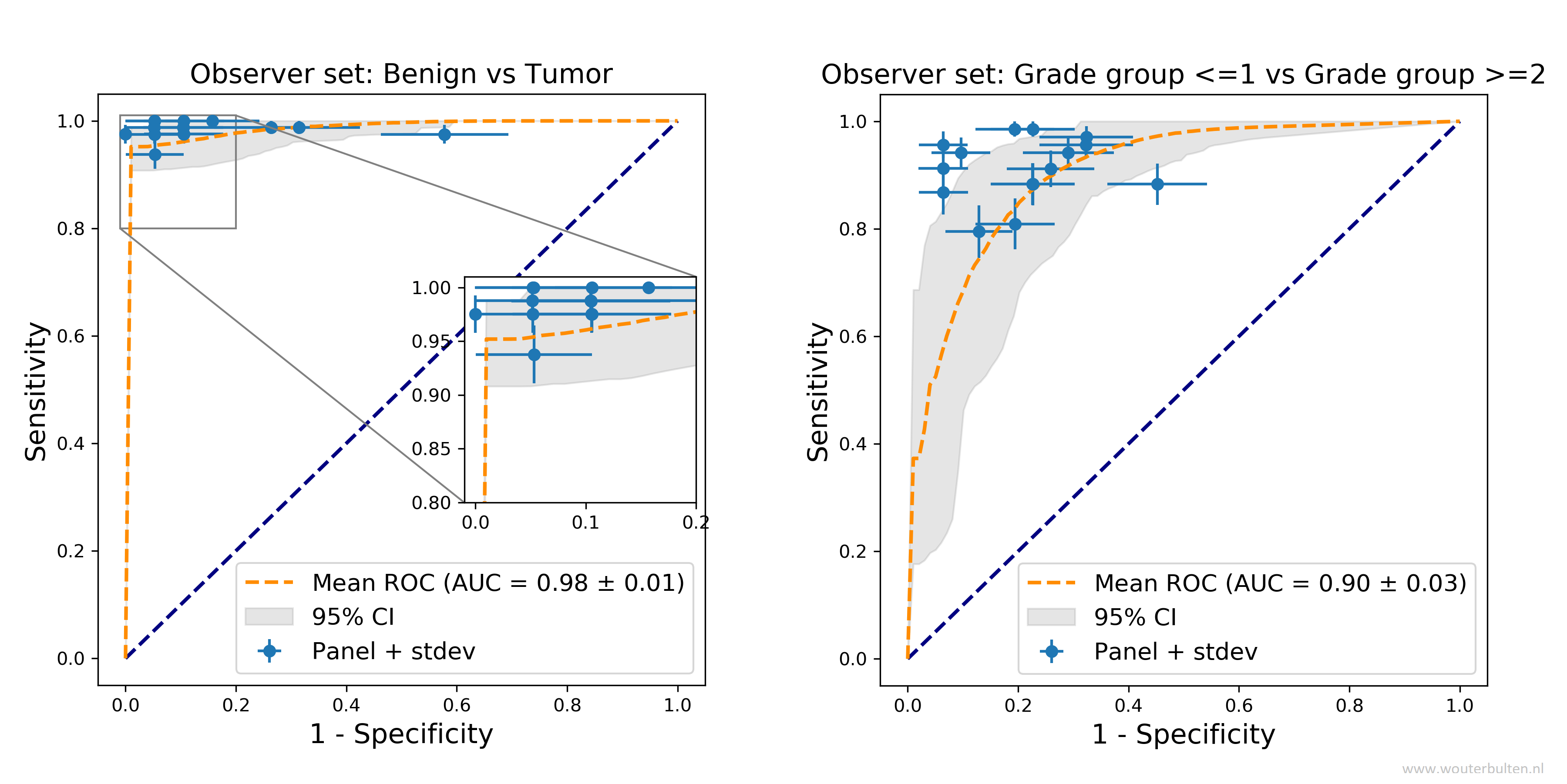
Comparison of the deep learning system with a panel of pathologists on two relevant patient risk groups.
Future challenges
There are still challenges to overcome before a system such as ours can be used in clinical practice. First of all, our system was developed and evaluated using data from a single center. Including data from different centers, with different stain protocols and scanners could increase the generalization ability of the system. Second, while the performance is high, it is not perfect, and the accuracy of the system could be improved. Though, at some point, it is difficult to say what is better given the high inter- and intra-rater agreement within prostate cancer grading.
Given our results, we conclude that there is clear use for automated systems for Gleason grading. Such systems can give feedback to a pathologist at expert levels, both in a first or second reader setting. AI systems and pathologists excel at different things, and, we firmly believe that a union of both has the most potential for the individual prostate cancer patient.
Interested in how this AI system can help pathologists? We have published follow-up research on how our AI system can assist pathologist during grading of prostate biopsies.
Try out the algorithm online
Interested on testing our algorithm on your own data? The algorithm we developed is available to try out online, without any requirements on deep learning hardware.
The online Gleason grading algorithm consists of multiple steps to prepare and analyze the data:
- For each biopsy, a tissue mask will be generated by a deep learning-based tissue segmentation system.
- Based on the input and a small set of target files, a CycleGAN-based normalization algorithm is trained.
- The trained normalization algorithm is applied to all input biopsies to normalize the data.
- Each normalized biopsy is processed by the automated Gleason grading system. The output of the system consists of an overlay and a set of metrics for each biopsy. The metrics include the biopsy-level grade group, the Gleason score, and predicted volume percentages.
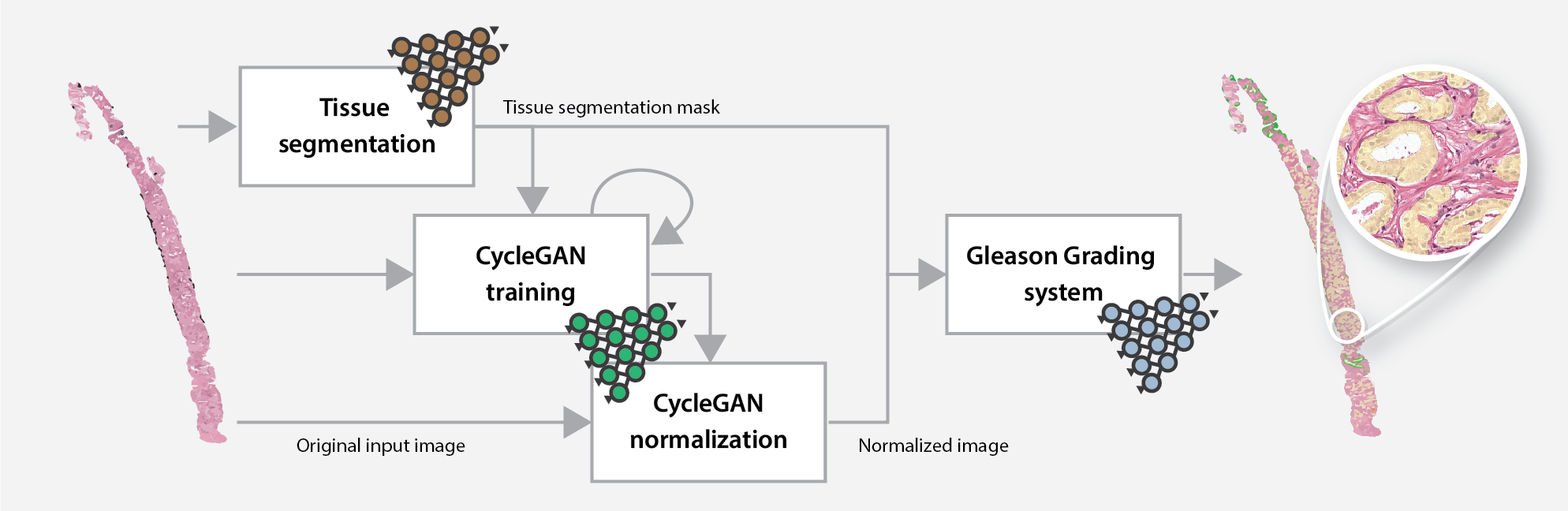
Overview of the online Gleason grading algorithm, including the normalization step.
More info
Download paper Download preprint
- More details can be found in the article at Lancet Oncology or in the preprint. Both the article and the preprint have supplementary materials with more details.
- A description of the tumor detection algorithm can be found in the paper on tumor detection.
- The epithelium segmentation system is described in a separate paper. The dataset is public, can be found online through Zenodo and is described in a blog post.
- This research was performed as part of the Computational Pathology Group and the Diagnostic Image Analysis Group of Radboud University Medical Center.
Please use the following to refer to our paper or this post:
Bulten, Wouter; Pinckaers, Hans; van Boven, Hester; Vink, Robert; de Bel, Thomas; van Ginneken, Bram; van der Laak, Jeroen; Hulsbergen-van de Kaa, Christina; Litjens, Geert, “Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study.” The Lancet Oncology (2020).
Or, if you prefer BibTeX:
@article{bulten2020automated,
title={Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study},
author={Bulten, Wouter and Pinckaers, Hans and van Boven, Hester and Vink, Robert and de Bel, Thomas and van Ginneken, Bram and van der Laak, Jeroen and Hulsbergen-van de Kaa, Christina and Litjens, Geert},
journal={The Lancet Oncology},
year={2020},
publisher={Elsevier}
}
Acknowledgements
This work was financed by a grant from the Dutch Cancer Society (KWF). We would also like to thank all pathologists that contributed to the observer experiment.
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
AI for diagnosis of prostate cancer: the PANDA challenge
Artificial intelligence (AI) for prostate cancer analysis is ready for clinical implementation, shows a global programming competition, the PANDA challenge.
Improve prostate cancer diagnosis: participate in the PANDA Gleason grading challenge
Can you build a deep learning model that can accurately grade protate biopsies? Participate in the PANDA challenge
The potential of AI in medicine: AI-assistance improves prostate cancer grading
In a completely new study we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.


















Waqas Majeed on