Within medicine, there is somewhat of an “AI hype.” Mainly driven by advances in deep learning, more and more studies show the potential of using artificial intelligence as a diagnostic tool. The same trend is present in the subfield I am working in. The field of pathology slowly transitions from being centered around microscopes to assessing tissue using a computer screen (i.e., digital pathology). With this change, computational models assisting the pathologist became within reach. A new field was born: computational pathology.
In recent years many studies have shown that deep learning-based tools can replicate histological tasks. For prostate cancer, the main topic of my Ph.D. project, detection models12 have been around for some time and are slowly becoming available to the clinic.3 The next step is building AI systems that can perform prognostic tasks, such as grading cancer. The first steps have already been made in this direction. We recently published our work on AI-based grading for prostate cancer in Lancet Oncology.4 In the same issue, colleagues from Karolinska (Sweden) showed similar findings.5 These studies, and others,6 showed that AI systems can achieve pathologist-level performance, or even outperform pathologists (within the limits of the study setup).
Regardless of these results, the question remains how much benefit these systems actually offer to individual patients? Does outperforming pathologists, also mean that it improves the diagnostic process? Despite the merits of deep learning systems, they are also constrained by limitations that can have dramatic effects on the output of such systems. The presence of artifacts, which are very common in the field of pathology, can drastically alter a prediction of a deep learning system. A human observer would not be hindered by, for example, ink on slides or cutting artifacts, and at least flag the case as ungradable. A deep learning system without countermeasures against such artifacts could happily predict high-grade cancer, even if the tissue is benign.
In the end, AI algorithms be applied in the clinic and outside of a controlled research setting. In such applications a pathologist will be in the loop that actively uses the system, or, in the event of a fully automated system, has to sign-off on the cases. In other medical domains, such as radiology, this kind of integration has already been investigated.7 Interestingly, within pathology, research on pathologists actually using AI systems is limited. Moreover, most studies focus on the benefits of computer-aided detection, and not on prognostic measures such as grading cancer.
After completing our study on automated Gleason grading for prostate cancer, we were curious to what extend our system could improve the diagnostic process. In a completely new study, now available Open Acccess through Modern Pathology, we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
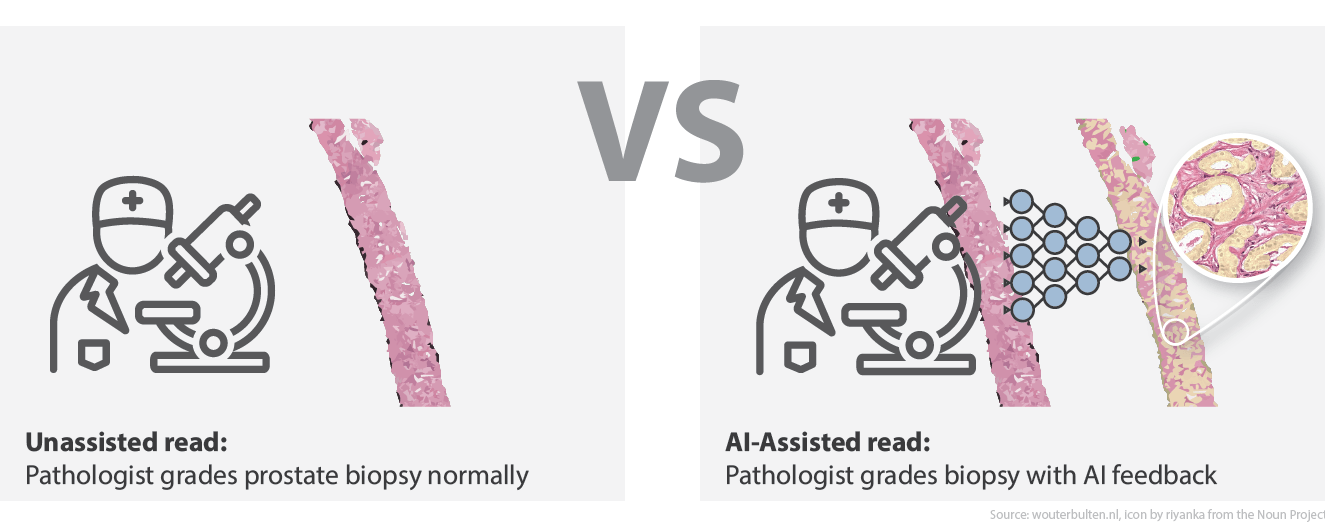
AI assistance for pathologists
When we developed the deep learning system, one of our design focusses was that the system should give interpretable output. Knowing the final diagnosis for a case is interesting, but it is more useful if a system can also show on what it based its decision. There are several ways to show output of a deep learning system8. In our case, we do this by letting the system highlight prostate glands that it finds malignant. Each detected malignant gland is marked either yellow ☗, orange ☗, or red ☗. This would make the inspection of the system’s prediction easy and should benefit the final diagnose, so we assumed.
In our Lancet Oncology publication,4 we showed that our deep learning system outperformed the manjority of the panel members. We, however, only compared the AI system to the panel. To investigate the integration of the AI system in the grading workflow of the pathologist, we invited the same panel to participate in a follow-up experiment. In total, 14 pathologists and residents joined our new study. The new experiment consisted of two reads: 1) the initial unassisted read as part of the Lancet Oncology publication, and 2) an AI-assisted read which occurred after a wash-out period.
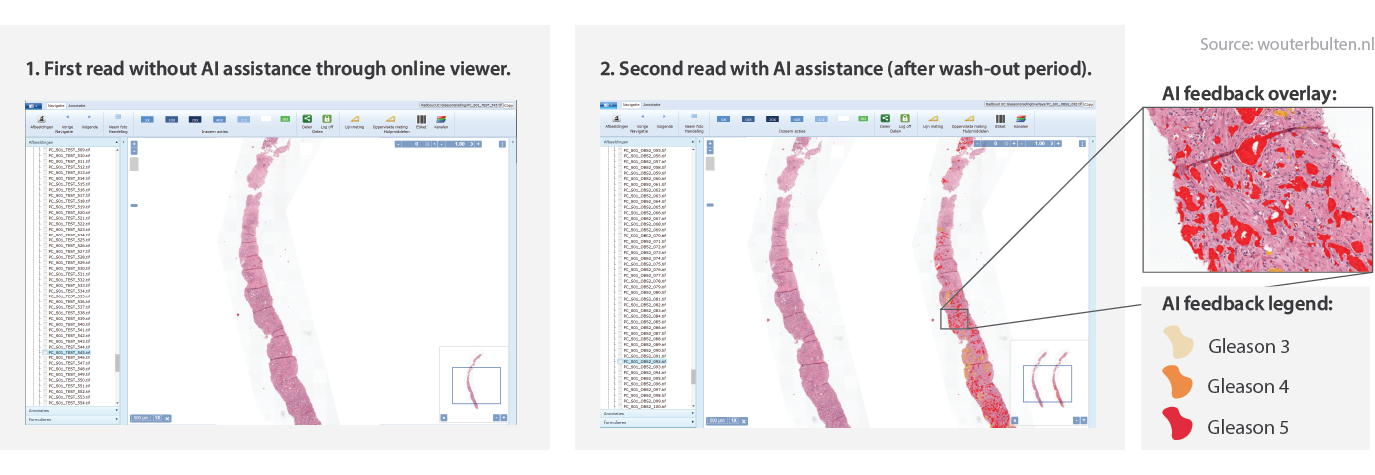
Overview of the study design. In the first read, the panel graded all biopsies without assistance. In the second read, AI assistance was presented next to the biopsy.
The unassisted read was already performed as part of the previous study. For the assisted read, we showed our panel of pathologists a set of 160 biopsies through an online viewer. Of these biopsies, 100 were part of the previous study; this way, we could compare performance between assisted and unassisted. The minimal time between reads was three months. The remaining 60 cases were used as controls.
For the pathologists, the difference between the two reads was the AI feedback. While in the unassisted read, panel members graded cases as they would normally. In the assisted read, we showed the output of the AI system next to the original biopsy (see figure, and live example). Additionally, the biopsy-level prediction of the system was also available to the panel (grade group and Gleason score).
For all of these biopsies, the reference standard was set by three uropathologists in consensus. Panel members determined the grade group of the biopsy, which we later compared to the reference standard.
Better performance, lower variability
After the assisted read, the scores of the individual readers were compared across the two runs. As the primary metric, we used quadratically weighted Cohen’s kappa. Whereas in the unassisted read, the panel achieved a median score of 0.79, this increased to 0.87 in the AI-assisted read (an increase of almost 10%). Besides this increase, the variability of the panel members dropped; the interquartile range of the panel’s kappa values dropped from 0.11 to 0.07.
Without assistance, the AI system outperformed 10 out of 14 observers (71%). In the assisted read, this flipped, and 9 out of 14 panel members exceeded the AI. On a group level, the AI-assisted pathologists outperformed not only the unassisted reads but also the AI itself.
Boxplot of the unassisted and AI-assisted read. In the unassited read (left), the performance of the system is higher than the median of the panel. In the assisted read (right), the panel outperforms both itself (unassisted) and the AI system.
When split out on experience level, less experienced pathologists benefitted the most. Most of the pathologists who had more than 15 years of experience, already scored similar or better than the AI system and had less to gain in terms of diagnostic performance. Still, the AI could have helped them grade the cases faster, something we did not explicitly test.
External validation
After the experiment on the internal dataset, we performed an additional experiment to test our hypothesis on external data. It is well known that the performance of deep learning systems can degrade when applied to unseen data. Additionally, by repeating our experiment, we were interested to see if the measured effects could be reproduced.
For this external validation, we made use of the Imagebase dataset,9 which consisted of 87 cases graded by 24 international experts in prostate pathology. The deep learning system was applied as-is to the dataset without any normalization of the data. The experiment was repeated in the same fashion: first an unassisted read, and, after a washout period, an assisted read with the AI predictions shown next to the original biopsy.
Of the 14 panel members, 12 joined the second experiment using the Imagebase dataset. In the unassisted read, the median pairwise agreement with the Imagebase panel was 0.73 (quadratically weighted Cohen’s kappa) and 9 out of the 11 panel members (75%) scored lower than the AI system. With AI assistance, the agreement increased significantly to 0.78 (p = 0.003), with the majority of the panel now outperforming the standalone AI system (10 out of 12, 83%). In contrast to the internal experiment, improvements could be seen for all but one of the panel members, with no clear effect of experience level.
Looking beyond diagnostic performance
Besides diagnostic performance, other factors are essential to increase the adoption of AI techniques in the diagnostic process10. Pathologists are often time-constrained and have high workloads. Any algorithm has to be easy to use and ideally speed up diagnosis. More research is welcome on how we can efficiently embed these new techniques in routine practice, especially in a field that is used to analog examination through microscopes.
In our study, we did not quantitatively measure the use of our system. Though, we did ask all panel members to fill in a survey regarding the grading process and their usage of the system. A summary of some of the findings:
- The majority of pathologists indicated that AI assistance sped up grading. It would be interesting to investigate the source of this time gain and to measure it quantitatively. Does it speed up finding the tumor? Makes it assessing tumor volumes faster?
- The gland-level prediction of the system was found most useful. The case-level label the least. This suggests that there is an added benefit of showing AI feedback on the source level. More detailed feedback also allows for more interaction between AI systems and pathologists.
- The majority of panel members would like to use an AI system during clinical practice.
For the other survey questions and the pathologist’s responses, please see the full paper.
Limitations, conclusion and future outlook
With our research, we aimed to set a new (small) step towards the clinical adoption of AI systems within pathology. For prostate cancer specifically, we showed that AI assistance reduced the observer variability of Gleason grading. Something that is highly desirable, as it could result in a stronger prognostic marker for individual patients and reduces the effect of the diagnosing pathologist on potential treatment decisions.
Of course, there are limitations to our study that open up many avenues of future research. For example, in our main experiment, we tested only two reads with a fixed order. While we were able to reproduce the same effect in a second experiment, additional research with larger datasets is welcome. Specifically, it would be exciting to investigate the effect of AI assistance over a more extended period. Maybe the benefit of AI assistance wears out over time, as pathologists learn from the feedback and apply this knowledge to unassisted reads. It is also plausible that pathologists become faster as they get accustomed to the feedback of the system. Ultimately, systems such as ours should be evaluated on a patient-level, which allows for new approaches such as automatically prioritizing slides.
In any case, the future is exciting, both from the AI perspective as well as on the implementation side. The tools are there to improve cancer diagnosis even further.
Example of AI feedback
Acknowledgements
Author list: Wouter Bulten, Maschenka Balkenhol, Jean-Joël Awoumou Belinga, Américo Brilhante, Aslı Çakır, Lars Egevad, Martin Eklund, Xavier Farré, Katerina Geronatsiou, Vincent Molinié, Guilherme Pereira, Paromita Roy, Günter Saile, Paulo Salles, Ewout Schaafsma, Joëlle Tschui, Anne-Marie Vos, ISUP Pathology Imagebase Expert Panel, Hester van Boven, Robert Vink, Jeroen van der Laak, Christina Hulsbergen-van der Kaa & Geert Litjens
This work was financed by a grant from the Dutch Cancer Society (KWF).
Please use the following to refer to our paper or this post:
Bulten, W., Balkenhol, M., Belinga, J.A. et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod Pathol (2020). https://doi.org/10.1038/s41379-020-0640-y
Or, if you prefer BibTeX:
@article{bulten2020artificial,
author={Bulten, Wouter and Balkenhol, Maschenka and Belinga, Jean-Jo{\"e}l Awoumou and Brilhante, Am{\'e}rico and {\c{C}}ak{\i}r, Asl{\i} and Egevad, Lars and Eklund, Martin and Farr{\'e}, Xavier and Geronatsiou, Katerina and Molini{\'e}, Vincent and Pereira, Guilherme and Roy, Paromita and Saile, G{\"u}nter and Salles, Paulo and Schaafsma, Ewout and Tschui, Jo{\"e}lle and Vos, Anne-Marie and Delahunt, Brett and Samaratunga, Hemamali and Grignon, David J. and Evans, Andrew J. and Berney, Daniel M. and Pan, Chin-Chen and Kristiansen, Glen and Kench, James G. and Oxley, Jon and Leite, Katia R. M. and McKenney, Jesse K. and Humphrey, Peter A. and Fine, Samson W. and Tsuzuki, Toyonori and Varma, Murali and Zhou, Ming and Comperat, Eva and Bostwick, David G. and Iczkowski, Kenneth A. and Magi-Galluzzi, Cristina and Srigley, John R. and Takahashi, Hiroyuki and van der Kwast, Theo and van Boven, Hester and Vink, Robert and van der Laak, Jeroen and Hulsbergen-van der Kaa, Christina and Litjens, Geert},
title={Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists},
journal={Modern Pathology},
year={2020},
month={Aug},
day={05},
abstract={The Gleason score is the most important prognostic marker for prostate cancer patients, but it suffers from significant observer variability. Artificial intelligence (AI) systems based on deep learning can achieve pathologist-level performance at Gleason grading. However, the performance of such systems can degrade in the presence of artifacts, foreign tissue, or other anomalies. Pathologists integrating their expertise with feedback from an AI system could result in a synergy that outperforms both the individual pathologist and the system. Despite the hype around AI assistance, existing literature on this topic within the pathology domain is limited. We investigated the value of AI assistance for grading prostate biopsies. A panel of 14 observers graded 160 biopsies with and without AI assistance. Using AI, the agreement of the panel with an expert reference standard increased significantly (quadratically weighted Cohen's kappa, 0.799 vs. 0.872; p{\thinspace}={\thinspace}0.019). On an external validation set of 87 cases, the panel showed a significant increase in agreement with a panel of international experts in prostate pathology (quadratically weighted Cohen's kappa, 0.733 vs. 0.786; p{\thinspace}={\thinspace}0.003). In both experiments, on a group-level, AI-assisted pathologists outperformed the unassisted pathologists and the standalone AI system. Our results show the potential of AI systems for Gleason grading, but more importantly, show the benefits of pathologist-AI synergy.},
issn={1530-0285},
doi={10.1038/s41379-020-0640-y},
url={https://doi.org/10.1038/s41379-020-0640-y}
}
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
AI for diagnosis of prostate cancer: the PANDA challenge
Artificial intelligence (AI) for prostate cancer analysis is ready for clinical implementation, shows a global programming competition, the PANDA challenge.
Improve prostate cancer diagnosis: participate in the PANDA Gleason grading challenge
Can you build a deep learning model that can accurately grade protate biopsies? Participate in the PANDA challenge
Epithelium segmentation using deep learning and immunohistochemistry
We developed a new deep learning method to segment epithelial tissue in digitized hematoxylin and eosin (H&E) stained prostatectomy slides using immunohistochemistry (IHC) as reference standard.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.
References
Litjens, G. et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286, Read online (2016). ↩︎
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301-1309, Read online (2019). ↩︎
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. The Lancet Oncology 21, 233-241, Read online (2020). ↩︎ ↩︎
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. The Lancet Oncology 21, 222-232, Read online (2020). ↩︎
Nagpal, K. et al. Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer. npj Digital Medicine, Read online (2018). ↩︎
Rodríguez-Ruiz, A. et al. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology 290, 305-314, Read online (2018). ↩︎
Interesting blog post on different ways of displaying model output: Communicating Model Uncertainty Over Space - How can we show a pathologist an AI model’s predictions ↩︎
Egevad L, Delahunt B, Berney DM, Bostwick DG, Cheville J, Comperat E, et al. Utility of pathology imagebase for standardisation of prostate cancer grading. Histopathology. 2018;73:8–18. Read online ↩︎
Cai CJ, Winter S, Steiner D, Wilcox L, Terry M. Hello AI: Uncovering the Onboarding Needs of Medical Practitioners for Human-AI Collaborative Decision-Making. Proceedings of the ACM on Human-Computer Interaction 2019;3:104. Read online ↩︎
